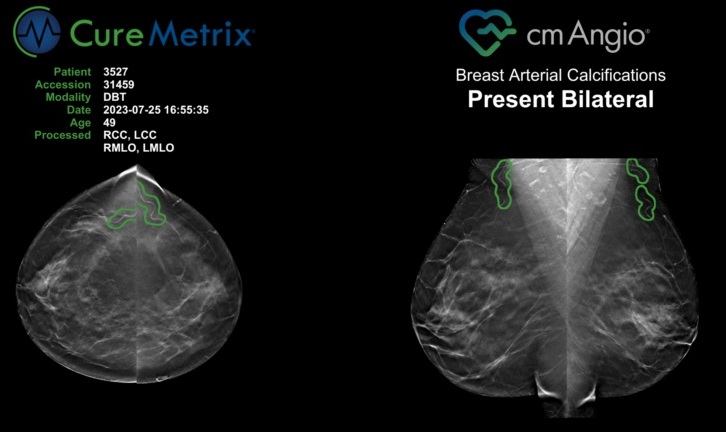
Expo
view channel
view channel
view channel
view channel
view channel
view channel
MRIUltrasoundNuclear MedicineGeneral/Advanced ImagingImaging ITIndustry News
Events

- AI Detects Early Signs of Aging from Chest X-Rays
- X-Ray Breakthrough Captures Three Image-Contrast Types in Single Shot
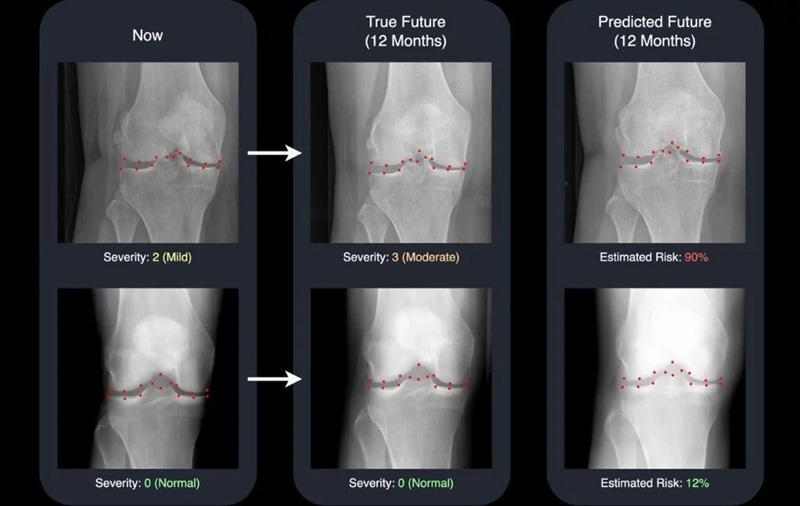
- AI Generates Future Knee X-Rays to Predict Osteoarthritis Progression Risk
- AI Algorithm Uses Mammograms to Accurately Predict Cardiovascular Risk in Women
- AI Hybrid Strategy Improves Mammogram Interpretation
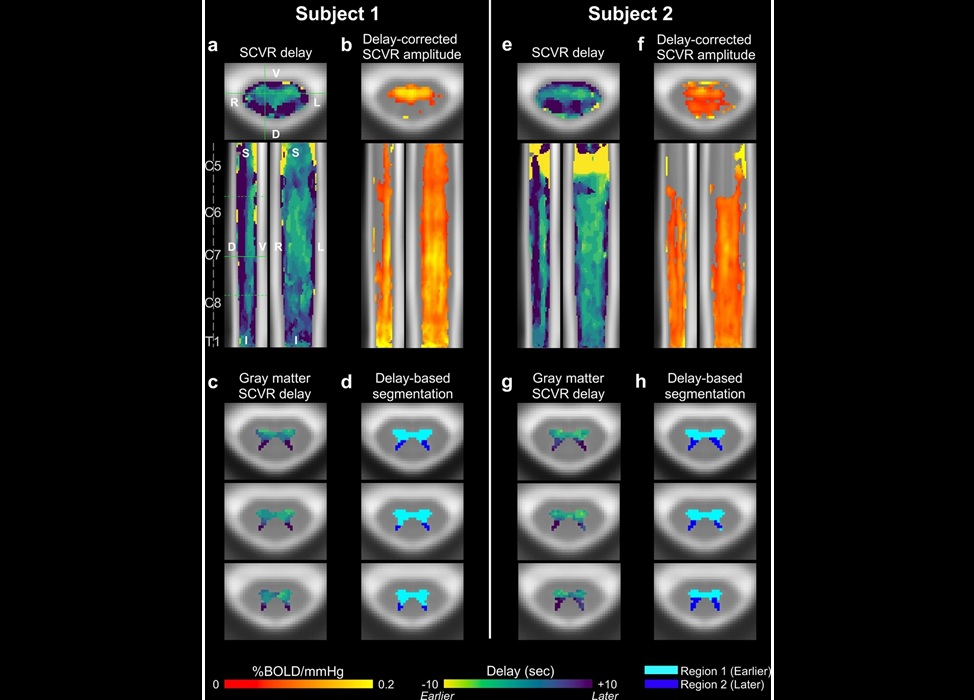
- Novel Imaging Approach to Improve Treatment for Spinal Cord Injuries
- AI-Assisted Model Enhances MRI Heart Scans
- AI Model Outperforms Doctors at Identifying Patients Most At-Risk of Cardiac Arrest
- New MRI Technique Reveals Hidden Heart Issues
- Shorter MRI Exam Effectively Detects Cancer in Dense Breasts
- Cancer “Flashlight” Shows Who Can Benefit from Targeted Treatments
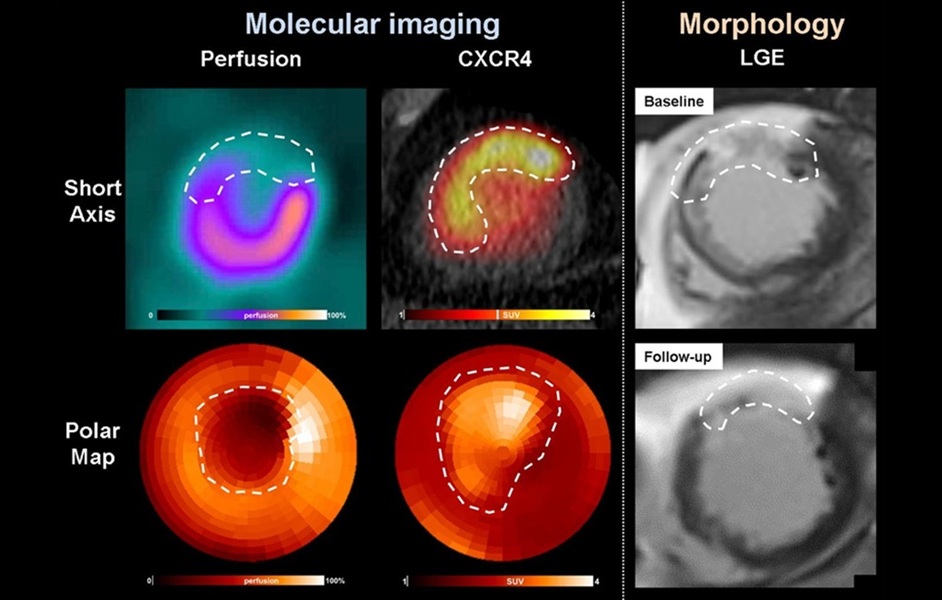
- PET Imaging of Inflammation Predicts Recovery and Guides Therapy After Heart Attack
- Radiotheranostic Approach Detects, Kills and Reprograms Aggressive Cancers
- New Imaging Solution Improves Survival for Patients with Recurring Prostate Cancer
- PET Tracer Enables Same-Day Imaging of Triple-Negative Breast and Urothelial Cancers
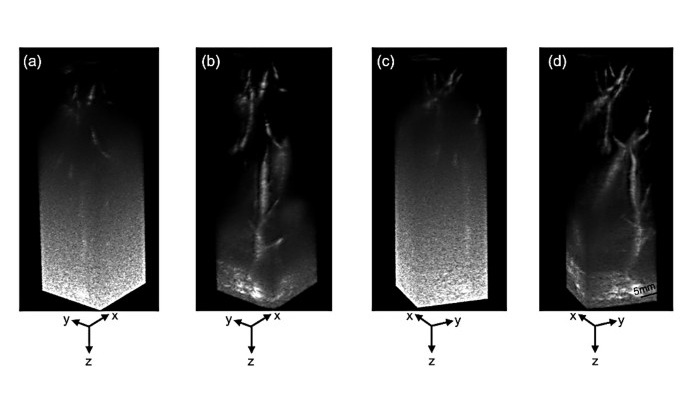
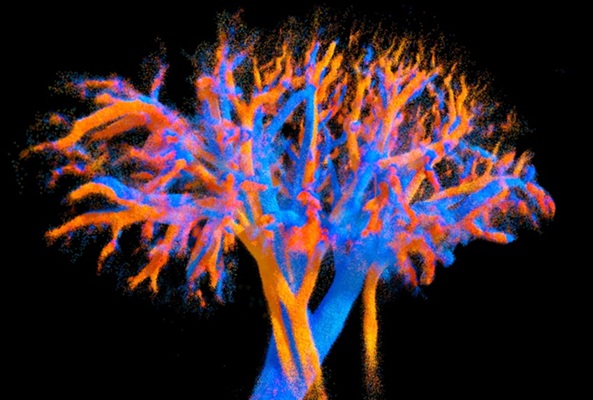
- Ultrasound Technique Visualizes Deep Blood Vessels in 3D Without Contrast Agents
- Ultrasound Probe Images Entire Organ in 4D
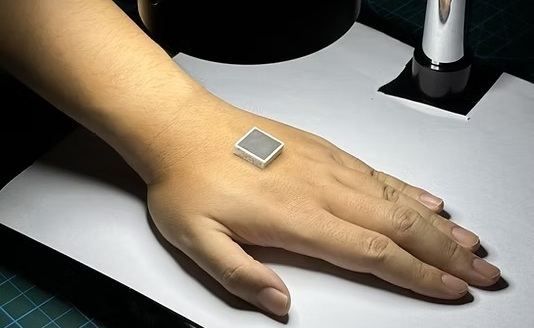
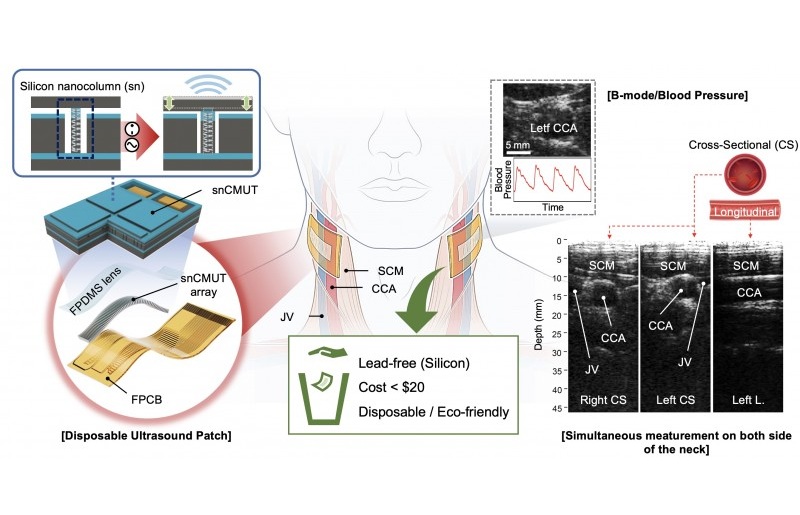
- Disposable Ultrasound Patch Performs Better Than Existing Devices
- Non-Invasive Ultrasound-Based Tool Accurately Detects Infant Meningitis
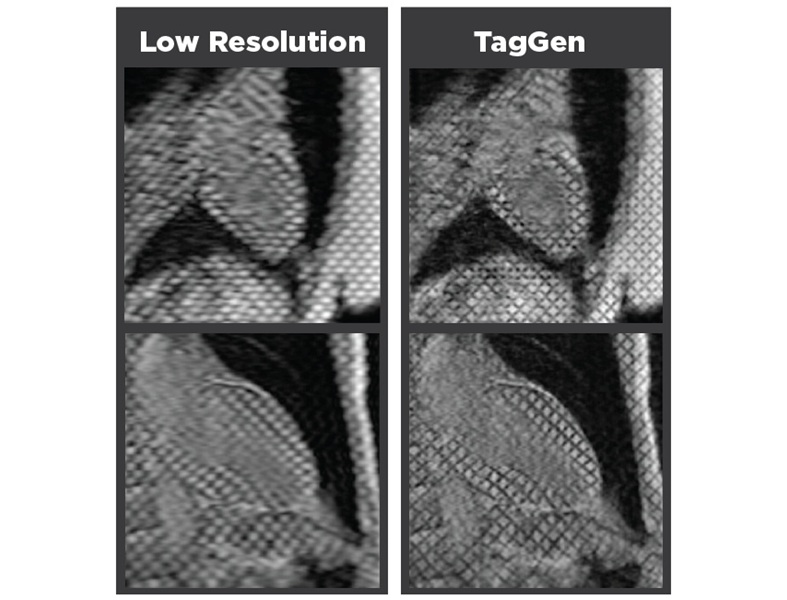
- Breakthrough Deep Learning Model Enhances Handheld 3D Medical Imaging
- Global AI in Medical Diagnostics Market to Be Driven by Demand for Image Recognition in Radiology
- AI-Based Mammography Triage Software Helps Dramatically Improve Interpretation Process
- Artificial Intelligence (AI) Program Accurately Predicts Lung Cancer Risk from CT Images
- Image Management Platform Streamlines Treatment Plans
- AI Technology for Detecting Breast Cancer Receives CE Mark Approval
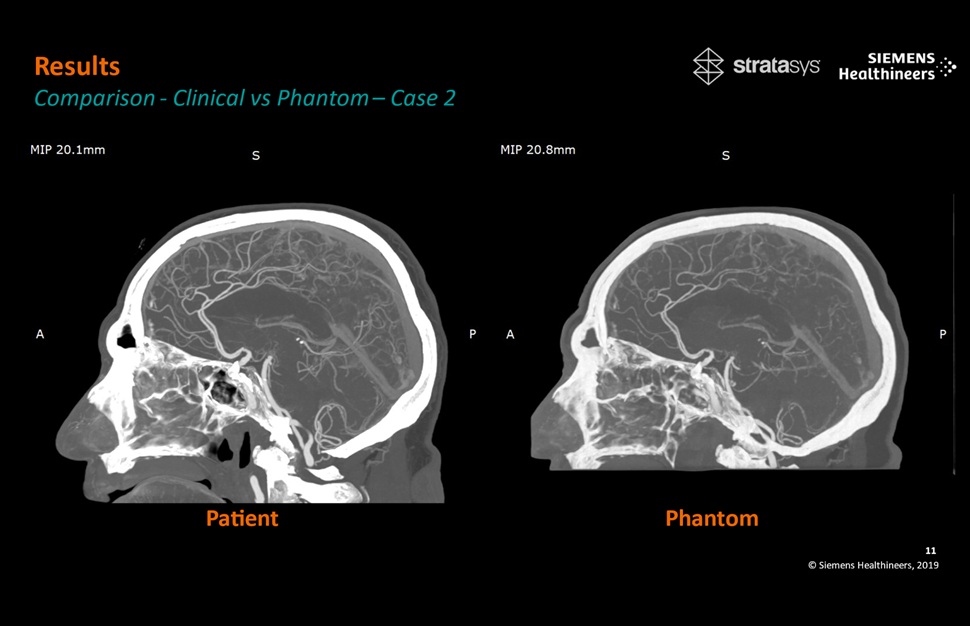
- Patient-Specific 3D-Printed Phantoms Transform CT Imaging
- Siemens and Sectra Collaborate on Enhancing Radiology Workflows
- Bracco Diagnostics and ColoWatch Partner to Expand Availability CRC Screening Tests Using Virtual Colonoscopy
- Mindray Partners with TeleRay to Streamline Ultrasound Delivery
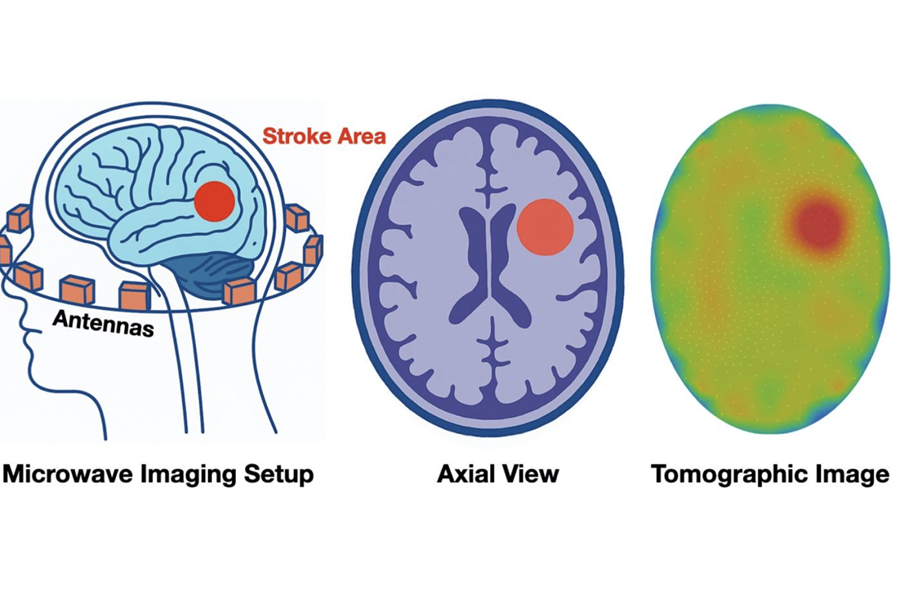
- Philips and Medtronic Partner on Stroke Care

 Expo
Expo
- AI Detects Early Signs of Aging from Chest X-Rays
- X-Ray Breakthrough Captures Three Image-Contrast Types in Single Shot
- AI Generates Future Knee X-Rays to Predict Osteoarthritis Progression Risk
- AI Algorithm Uses Mammograms to Accurately Predict Cardiovascular Risk in Women
- AI Hybrid Strategy Improves Mammogram Interpretation
- Novel Imaging Approach to Improve Treatment for Spinal Cord Injuries
- AI-Assisted Model Enhances MRI Heart Scans
- AI Model Outperforms Doctors at Identifying Patients Most At-Risk of Cardiac Arrest
- New MRI Technique Reveals Hidden Heart Issues
- Shorter MRI Exam Effectively Detects Cancer in Dense Breasts
- Cancer “Flashlight” Shows Who Can Benefit from Targeted Treatments
- PET Imaging of Inflammation Predicts Recovery and Guides Therapy After Heart Attack
- Radiotheranostic Approach Detects, Kills and Reprograms Aggressive Cancers
- New Imaging Solution Improves Survival for Patients with Recurring Prostate Cancer
- PET Tracer Enables Same-Day Imaging of Triple-Negative Breast and Urothelial Cancers
- Ultrasound Technique Visualizes Deep Blood Vessels in 3D Without Contrast Agents
- Ultrasound Probe Images Entire Organ in 4D
- Disposable Ultrasound Patch Performs Better Than Existing Devices
- Non-Invasive Ultrasound-Based Tool Accurately Detects Infant Meningitis
- Breakthrough Deep Learning Model Enhances Handheld 3D Medical Imaging
- Global AI in Medical Diagnostics Market to Be Driven by Demand for Image Recognition in Radiology
- AI-Based Mammography Triage Software Helps Dramatically Improve Interpretation Process
- Artificial Intelligence (AI) Program Accurately Predicts Lung Cancer Risk from CT Images
- Image Management Platform Streamlines Treatment Plans
- AI Technology for Detecting Breast Cancer Receives CE Mark Approval
- Patient-Specific 3D-Printed Phantoms Transform CT Imaging
- Siemens and Sectra Collaborate on Enhancing Radiology Workflows
- Bracco Diagnostics and ColoWatch Partner to Expand Availability CRC Screening Tests Using Virtual Colonoscopy
- Mindray Partners with TeleRay to Streamline Ultrasound Delivery
- Philips and Medtronic Partner on Stroke Care